Working with ASCII momentum tuple files#
Pawian usually imports its data from momentum tuples written to an ASCII text file. Each line consists of four values: the energy and the \(x\), \(y\), \(z\)-components of the 3-momentum. The lines are grouped by event and can be preceded by an event weight. An example of two weighted events of three particles each would be:
0.99407
-0.00357645 0.0962561 0.0181079 0.170545
0.224019 0.623156 0.215051 1.99057
-0.174404 -0.719412 -0.233159 2.0243
0.990748
-0.0328198 0.0524406 0.0310079 0.155783
-0.619592 0.141315 0.32135 1.99619
0.698477 -0.193756 -0.352357 2.03593
The pawian.data module imports such an ASCII file to a nicely formatted pandas.DataFrame and provides a few accessors that facilitate visualization of the content of the ASCII file.
The fact that we works with a pandas.DataFrame also allows one to make selections of the content and write the filtered data set to another ASCII file for Pawian (and whatever other format is already supported by pandas.DataFrame).
Import data#
In this example, we use the test files provided the pawian.data module folder in the repository.
The data file describes momentum tuples for a \(e^+e^- \to \pi+D^0D^+\) decay (in that order!). This information can be passed on to the read_ascii() function to create a pandas.DataFrame.
from pawian.data import read_ascii
particles = ["pi+", "D0", "D-"]
frame = read_ascii(filename_data, particles=particles)
frame
| Particle | pi+ | D0 | D- | weight | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Momentum | p_x | p_y | p_z | E | p_x | p_y | p_z | E | p_x | p_y | p_z | E | |
| 0 | -0.003576 | 0.096256 | 0.018108 | 0.170545 | 0.224019 | 0.623156 | 0.215051 | 1.99057 | -0.174404 | -0.719412 | -0.233159 | 2.02430 | 0.994070 |
| 1 | -0.032820 | 0.052441 | 0.031008 | 0.155783 | -0.619592 | 0.141315 | 0.321350 | 1.99619 | 0.698477 | -0.193756 | -0.352357 | 2.03593 | 0.990748 |
| 2 | 0.109609 | -0.192790 | 0.045853 | 0.266016 | -0.303136 | -0.204413 | -0.444309 | 1.95159 | 0.239557 | 0.397203 | 0.398456 | 1.96707 | 0.281660 |
| 3 | 0.050984 | -0.039164 | -0.041701 | 0.159223 | 0.627018 | -0.352803 | -0.081468 | 2.00047 | -0.632071 | 0.391967 | 0.123168 | 2.01588 | 0.992387 |
| 4 | 0.064191 | 0.002905 | -0.062571 | 0.165903 | 0.079182 | 0.241238 | -0.666256 | 1.99649 | -0.097326 | -0.244142 | 0.728827 | 2.02379 | 0.534049 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 995 | 0.208905 | 0.228950 | 0.060918 | 0.345326 | 0.011720 | -0.024639 | -0.443599 | 1.91707 | -0.174541 | -0.204311 | 0.382681 | 1.92720 | 0.179032 |
| 996 | 0.049370 | -0.053923 | 0.084898 | 0.178976 | 0.363975 | -0.294093 | -0.565002 | 2.00395 | -0.367383 | 0.348016 | 0.480105 | 1.99550 | 0.652028 |
| 997 | 0.014450 | -0.050924 | -0.038706 | 0.154208 | 0.420817 | -0.151503 | -0.588410 | 2.00596 | -0.389284 | 0.202427 | 0.627117 | 2.02021 | 0.997076 |
| 998 | -0.109077 | -0.041651 | 0.033446 | 0.185016 | -0.494181 | -0.402043 | 0.058469 | 1.97152 | 0.649313 | 0.443694 | -0.091916 | 2.03036 | 0.166129 |
| 999 | -0.048741 | -0.034371 | 0.017186 | 0.152749 | -0.282073 | -0.191036 | 0.642406 | 2.00159 | 0.376848 | 0.225407 | -0.659593 | 2.03060 | 0.986252 |
1000 rows × 13 columns
Investigate content of the data frame#
Notice that the data frame makes use of multi-indexing for the columns. This allows us for instance to make easy selections per particle, like this:
| Momentum | p_x | p_y | p_z | E |
|---|---|---|---|---|
| 0 | -0.177980 | -0.623156 | -0.215051 | 2.194845 |
| 1 | 0.665657 | -0.141315 | -0.321349 | 2.191713 |
| 2 | 0.349166 | 0.204413 | 0.444309 | 2.233086 |
| 3 | -0.581087 | 0.352803 | 0.081467 | 2.175103 |
| 4 | -0.033134 | -0.241237 | 0.666256 | 2.189693 |
| ... | ... | ... | ... | ... |
| 995 | 0.034364 | 0.024639 | 0.443599 | 2.272526 |
| 996 | -0.318013 | 0.294093 | 0.565003 | 2.174476 |
| 997 | -0.374834 | 0.151503 | 0.588411 | 2.174418 |
| 998 | 0.540236 | 0.402043 | -0.058469 | 2.215376 |
| 999 | 0.328107 | 0.191036 | -0.642407 | 2.183349 |
1000 rows × 4 columns
frame[["pi+", "D0"]].mean()
Particle Momentum
pi+ p_x 0.001480
p_y 0.006563
p_z -0.001542
E 0.207283
D0 p_x 0.035191
p_y 0.000136
p_z -0.008241
E 1.977187
dtype: float64
Even better, we immediately have all powerful techniques of a DataFrame at our disposal:
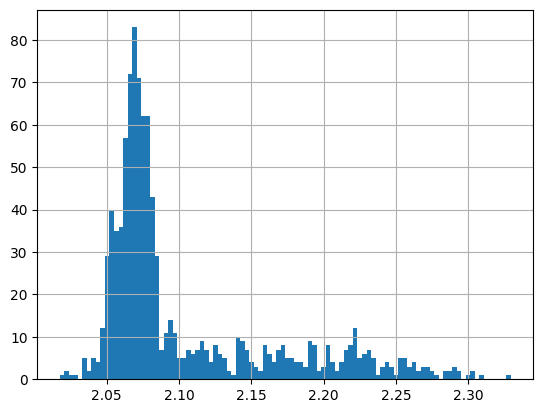
frame["D-"].hist(bins=50);
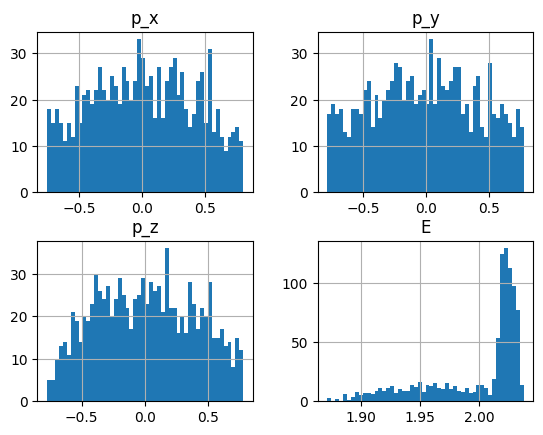
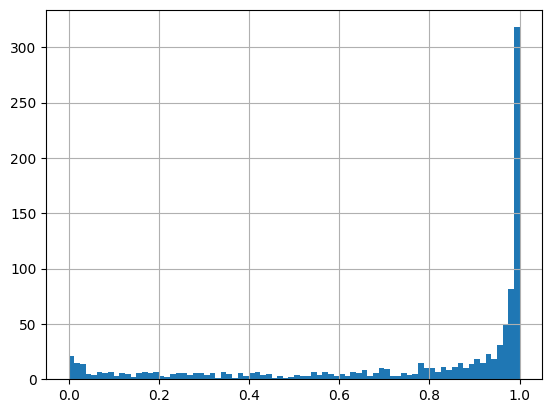
frame["weight"].hist(bins=80);
Special accessors#
Now that we have imported from the pawian.data sub-module, a few simple accessors to the data frame have become available in the namespace pwa of the DataFrame (see PwaAccessor). They can be called from the pwa namespace like so:
print("Has weights: ", frame.pwa.has_weights)
print("Contains particles:", frame.pwa.particles)
print("Contains momenta: ", frame.pwa.momentum_labels)
Has weights: True
Contains particles: ['pi+', 'D0', 'D-']
Contains momenta: ['p_x', 'p_y', 'p_z', 'E']
The accessors also allow to get kinematic variables:
| Particle | pi+ | D0 | D- | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Momentum | p_x | p_y | p_z | p_x | p_y | p_z | p_x | p_y | p_z |
| 0 | -0.003576 | 0.096256 | 0.018108 | 0.224019 | 0.623156 | 0.215051 | -0.174404 | -0.719412 | -0.233159 |
| 1 | -0.032820 | 0.052441 | 0.031008 | -0.619592 | 0.141315 | 0.321350 | 0.698477 | -0.193756 | -0.352357 |
| 2 | 0.109609 | -0.192790 | 0.045853 | -0.303136 | -0.204413 | -0.444309 | 0.239557 | 0.397203 | 0.398456 |
| 3 | 0.050984 | -0.039164 | -0.041701 | 0.627018 | -0.352803 | -0.081468 | -0.632071 | 0.391967 | 0.123168 |
| 4 | 0.064191 | 0.002905 | -0.062571 | 0.079182 | 0.241238 | -0.666256 | -0.097326 | -0.244142 | 0.728827 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 995 | 0.208905 | 0.228950 | 0.060918 | 0.011720 | -0.024639 | -0.443599 | -0.174541 | -0.204311 | 0.382681 |
| 996 | 0.049370 | -0.053923 | 0.084898 | 0.363975 | -0.294093 | -0.565002 | -0.367383 | 0.348016 | 0.480105 |
| 997 | 0.014450 | -0.050924 | -0.038706 | 0.420817 | -0.151503 | -0.588410 | -0.389284 | 0.202427 | 0.627117 |
| 998 | -0.109077 | -0.041651 | 0.033446 | -0.494181 | -0.402043 | 0.058469 | 0.649313 | 0.443694 | -0.091916 |
| 999 | -0.048741 | -0.034371 | 0.017186 | -0.282073 | -0.191036 | 0.642406 | 0.376848 | 0.225407 | -0.659593 |
1000 rows × 9 columns
frame.pwa.mass.mean()
Particle
D- 1.86961
D0 1.86484
pi+ 0.13957
dtype: float64
And the best part: you can just add the vectors and do analysis on for instance their combined invariant mass!
Selecting and exporting#
As mentioned, pandas.DataFrame allows us to make certain selections:
| Particle | pi+ | D0 | D- | weight | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Momentum | p_x | p_y | p_z | E | p_x | p_y | p_z | E | p_x | p_y | p_z | E | |
| 0 | -0.003576 | 0.096256 | 0.018108 | 0.170545 | 0.224019 | 0.623156 | 0.215051 | 1.99057 | -0.174404 | -0.719412 | -0.233159 | 2.02430 | 0.994070 |
| 1 | -0.032820 | 0.052441 | 0.031008 | 0.155783 | -0.619592 | 0.141315 | 0.321350 | 1.99619 | 0.698477 | -0.193756 | -0.352357 | 2.03593 | 0.990748 |
| 3 | 0.050984 | -0.039164 | -0.041701 | 0.159223 | 0.627018 | -0.352803 | -0.081468 | 2.00047 | -0.632071 | 0.391967 | 0.123168 | 2.01588 | 0.992387 |
| 6 | -0.033936 | 0.042558 | 0.050071 | 0.157955 | -0.591966 | 0.113080 | 0.343726 | 1.98972 | 0.671870 | -0.155639 | -0.393797 | 2.03129 | 0.992469 |
| 7 | -0.005202 | -0.042270 | -0.071584 | 0.162536 | -0.110162 | -0.045471 | -0.690215 | 1.99204 | 0.161362 | 0.087741 | 0.761799 | 2.02719 | 0.991051 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 991 | -0.018286 | -0.047475 | -0.044260 | 0.155006 | -0.557088 | -0.343712 | -0.265556 | 1.99415 | 0.621370 | 0.391187 | 0.309815 | 2.03238 | 0.991111 |
| 992 | -0.001543 | 0.079682 | -0.043369 | 0.166470 | 0.181800 | 0.656877 | -0.171703 | 1.99290 | -0.134226 | -0.736559 | 0.215072 | 2.02540 | 0.998521 |
| 994 | 0.058329 | 0.050061 | -0.017219 | 0.160265 | 0.354361 | 0.428801 | -0.460858 | 1.99986 | -0.366689 | -0.478862 | 0.478077 | 2.02182 | 0.998338 |
| 997 | 0.014450 | -0.050924 | -0.038706 | 0.154208 | 0.420817 | -0.151503 | -0.588410 | 2.00596 | -0.389284 | 0.202427 | 0.627117 | 2.02021 | 0.997076 |
| 999 | -0.048741 | -0.034371 | 0.017186 | 0.152749 | -0.282073 | -0.191036 | 0.642406 | 2.00159 | 0.376848 | 0.225407 | -0.659593 | 2.03060 | 0.986252 |
480 rows × 13 columns
The frame can then be exported to an ASCII file that can be parsed by pawian like so:
output_file = "selected_data.dat"
selection.pwa.write_ascii(output_file)
imported_frame = read_ascii(output_file, particles)
imported_frame
| Particle | pi+ | D0 | D- | weight | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Momentum | p_x | p_y | p_z | E | p_x | p_y | p_z | E | p_x | p_y | p_z | E | |
| 0 | -0.003576 | 0.096256 | 0.018108 | 0.170545 | 0.224019 | 0.623156 | 0.215051 | 1.99057 | -0.174404 | -0.719412 | -0.233159 | 2.02430 | 0.994070 |
| 1 | -0.032820 | 0.052441 | 0.031008 | 0.155783 | -0.619592 | 0.141315 | 0.321350 | 1.99619 | 0.698477 | -0.193756 | -0.352357 | 2.03593 | 0.990748 |
| 2 | 0.050984 | -0.039164 | -0.041701 | 0.159223 | 0.627018 | -0.352803 | -0.081468 | 2.00047 | -0.632071 | 0.391967 | 0.123168 | 2.01588 | 0.992387 |
| 3 | -0.033936 | 0.042558 | 0.050071 | 0.157955 | -0.591966 | 0.113080 | 0.343726 | 1.98972 | 0.671870 | -0.155639 | -0.393797 | 2.03129 | 0.992469 |
| 4 | -0.005202 | -0.042270 | -0.071584 | 0.162536 | -0.110162 | -0.045471 | -0.690215 | 1.99204 | 0.161362 | 0.087741 | 0.761799 | 2.02719 | 0.991051 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 475 | -0.018286 | -0.047475 | -0.044260 | 0.155006 | -0.557088 | -0.343712 | -0.265556 | 1.99415 | 0.621370 | 0.391187 | 0.309815 | 2.03238 | 0.991111 |
| 476 | -0.001543 | 0.079682 | -0.043369 | 0.166470 | 0.181800 | 0.656877 | -0.171703 | 1.99290 | -0.134226 | -0.736559 | 0.215072 | 2.02540 | 0.998521 |
| 477 | 0.058329 | 0.050061 | -0.017219 | 0.160265 | 0.354361 | 0.428801 | -0.460858 | 1.99986 | -0.366689 | -0.478862 | 0.478077 | 2.02182 | 0.998338 |
| 478 | 0.014450 | -0.050924 | -0.038706 | 0.154208 | 0.420817 | -0.151503 | -0.588410 | 2.00596 | -0.389284 | 0.202427 | 0.627117 | 2.02021 | 0.997076 |
| 479 | -0.048741 | -0.034371 | 0.017186 | 0.152749 | -0.282073 | -0.191036 | 0.642406 | 2.00159 | 0.376848 | 0.225407 | -0.659593 | 2.03060 | 0.986252 |
480 rows × 13 columns